Other Resources
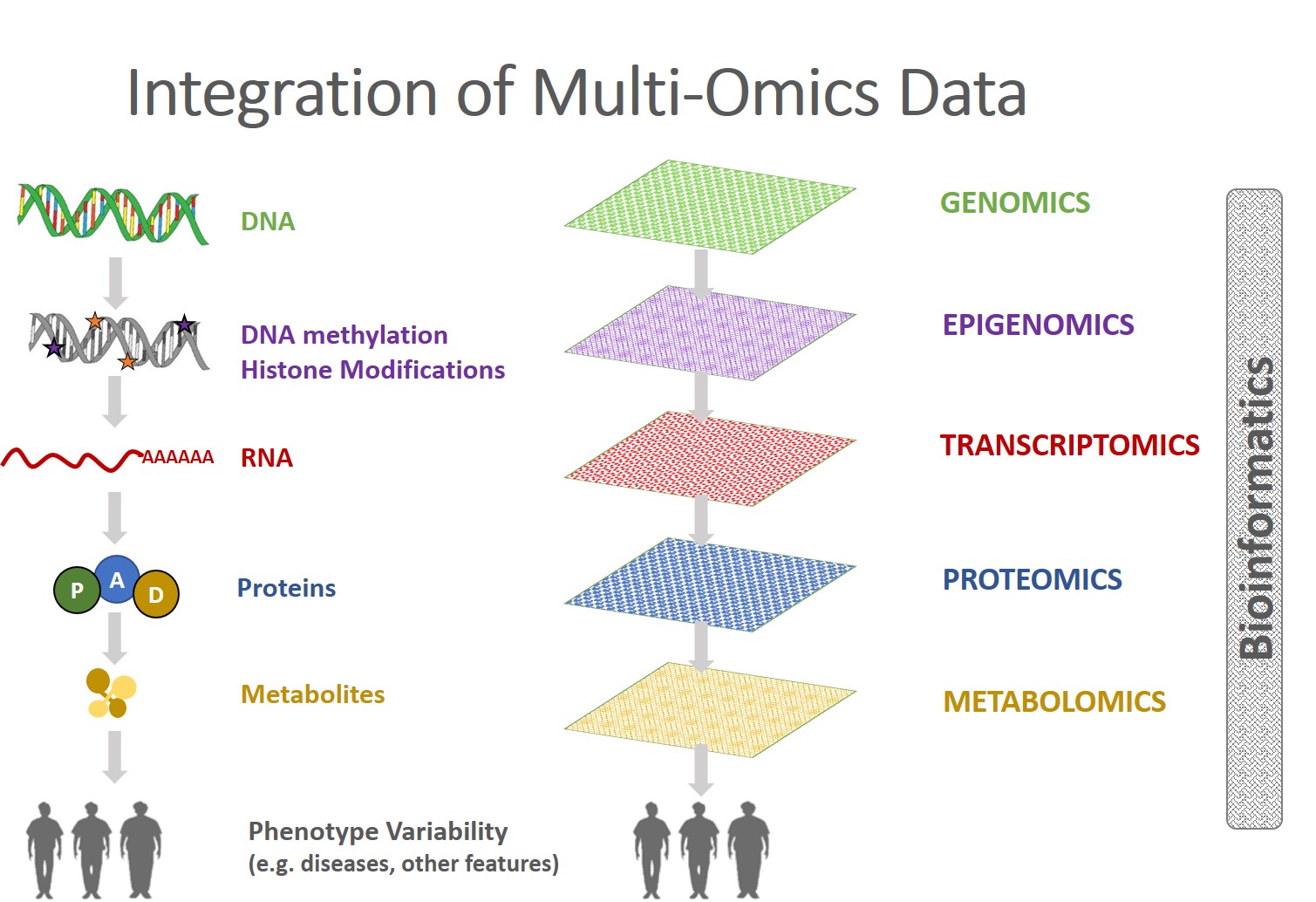
Table of Contents
Genome Databases



Multi-Omics Profiles

Genotype-Tissue Expression (GTEx) portal contains genotypes and transcriptome profiles of 53 non-diseased tissues from almost 1000 individuals, allwoing to study tissue-specific expression and regulations (eQTLs, sQTLs).



Multi-Omics Data
Raw data for all organisms, but requires analysis!


Drug Discovery


Other useful tools




